Plant Physiology 161: 1903-1917 (2013)
Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium DELAY OF GERMINATION1 Genes [W][OA]
School of Biological Sciences, Plant Molecular Science and Centre for Systems and Synthetic Biology, Royal Holloway, University of London, Egham, Surrey TW20 0EX, United Kingdom (KG, AV, GLM);
Web: 'The Seed Biology Place' - www.seedbiology.eu
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany/Plant Physiology, D-79104 Freiburg, Germany (KG, AV, ABM, GLM)
Universität Osnabrück, Fachbereich Biologie, Botanik, D-49069 Osnabrück, Germany (KS, KM)
Received December 24, 2012; Accepted Feburary 19, 2013; Published Feburary 20, 2013.
DOI:10.1104/pp.112.213298
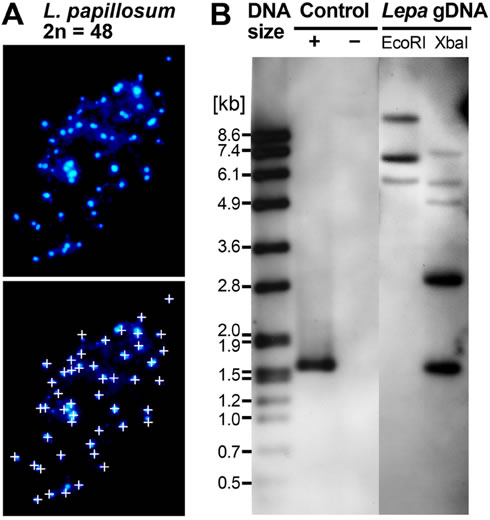
Figure 4. Chromosome number and DOG1 gene analysis of the Lepidium papillosum genome.
A, DAPI-stained mitotic chromosome spreads from flower bud tissue exhibiting 2n = 48 chromosomes. For better counting, chromosomes are labeled by small white crosses in the bottom image.
B, Southern analysis of L. papillosum genomic DNA (Lepa gDNA) digested with EcoRI or XbaI and hybridised with a 0.35 kb LesaDOG1 Exon 1 probe shows the presence of 3 to 5 hybridizing bands indicative for LepaDOG1 genes. Note that neither LesaDOG1, nor the cloned genomic LepaDOG1 fragments (Supplemental Table S1) contain EcoRI or XbaI restriction sites within the region covered by the Southern probe. Hybridization controls (CON) are plasmids with (+) or without (-) LesaDOG1 full-length genomic DNA inserts; left lane: DNA molecular mass ladder.
| Article in PDF format (2 MB) Supplementary data file (4.9 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |