Plant Physiology 161: 1903-1917 (2013)
Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium DELAY OF GERMINATION1 Genes [W][OA]
School of Biological Sciences, Plant Molecular Science and Centre for Systems and Synthetic Biology, Royal Holloway, University of London, Egham, Surrey TW20 0EX, United Kingdom (KG, AV, GLM);
Web: 'The Seed Biology Place' - www.seedbiology.eu
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany/Plant Physiology, D-79104 Freiburg, Germany (KG, AV, ABM, GLM)
Universität Osnabrück, Fachbereich Biologie, Botanik, D-49069 Osnabrück, Germany (KS, KM)
Received December 24, 2012; Accepted Feburary 19, 2013; Published Feburary 20, 2013.
DOI:10.1104/pp.112.213298
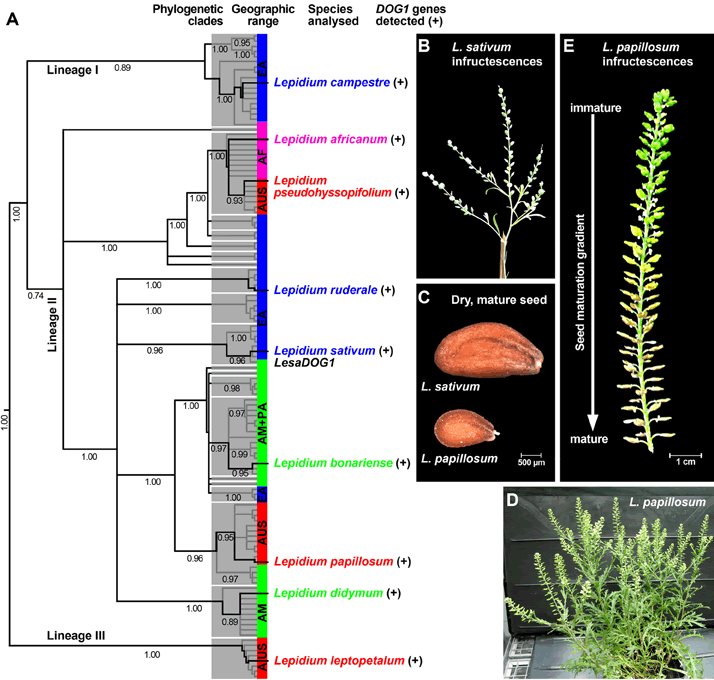
Figure 1. Phylogeny of the genus Lepidium, the distribution of the DOG1 gene, and infructescence morphology of Lepidium sativum and Lepidium papillosum.
A, Distribution of DOG1 homologs in representatives of all major lineages, subclades, and geographic ranges of Lepidium. Shown is the Bayesian 50% majority rule consensus tree based on ITS-sequence analysis, with mean posterior probability values (clade credibilities) below branches; phylogeny redrawn from Mummenhoff et al. (2009). For DOG1 gene sequences see Supplemental Table S1.
B-E, Comparison of infructescences and seeds of L. sativum and L. papillosum. Within the 'well-ordered' infructescences of L. papillosum the seed maturation gradient is accompanied by primary dormancy induction.
Abbreviations: AF, Africa; AM, America; AUS, Australia+New Zealand; EA, Eurasia; PA, Pacific Islands (Hawaii).
| Article in PDF format (2 MB) Supplementary data file (4.9 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |