Plant Physiology 155: 1851-1870 (2011)
Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: A global tissue specific transcript analysis
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany / Plant Physiology, Schänzlestr. 1, D-79104 Freiburg, Germany (A.L., Ke.M., K.O., G.L.-M.)
College of Life Sciences, South China Agricultural University, Guangzhou 510642, China (X.W.)
Department of Biological Sciences, Simon Fraser University, 8888 University Drive, Burnaby BC V5A 1S6, Canada (Ke.M.)
Received November 23, 2010; accepted February 7, 2011; published February 14, 2011
DOI 10.1104/pp.110.169706
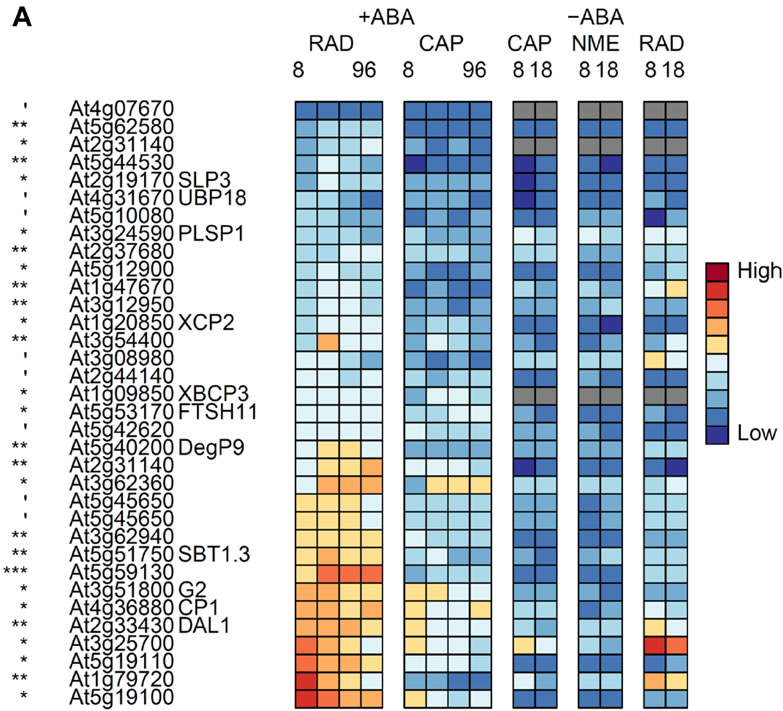
A,Genes in the TAGGIT protein degradation category that are up-regulated in the RAD.
B,Genes in the TAGGIT protein degradation category that are up-regulated in the CAP.
C, Genes encoding F box proteins that are significantly differentially expressed between the RAD and CAP.
D, Genes encoding RING finger E3 ligaseproteins that are significantly differentially expressed between the RAD and CAP .
', *, **, *** indicate that transcript numbers are significantly different between the tissues on +ABA-arrays at P<0.1, <0.05, <0.01, <0.001 respectively. Genes not present in the data sets are coloured grey.
Supplemental Figure S2. Heat maps showing the expression levels of genes encoding proteins associated with posttranslational modification.
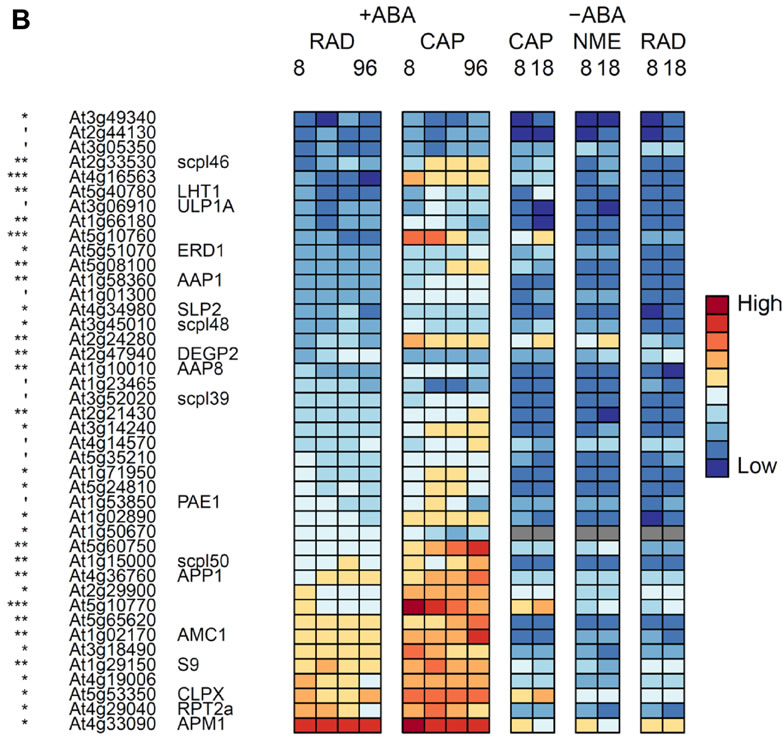
A,Genes in the TAGGIT protein degradation category that are up-regulated in the RAD.
B,Genes in the TAGGIT protein degradation category that are up-regulated in the CAP.
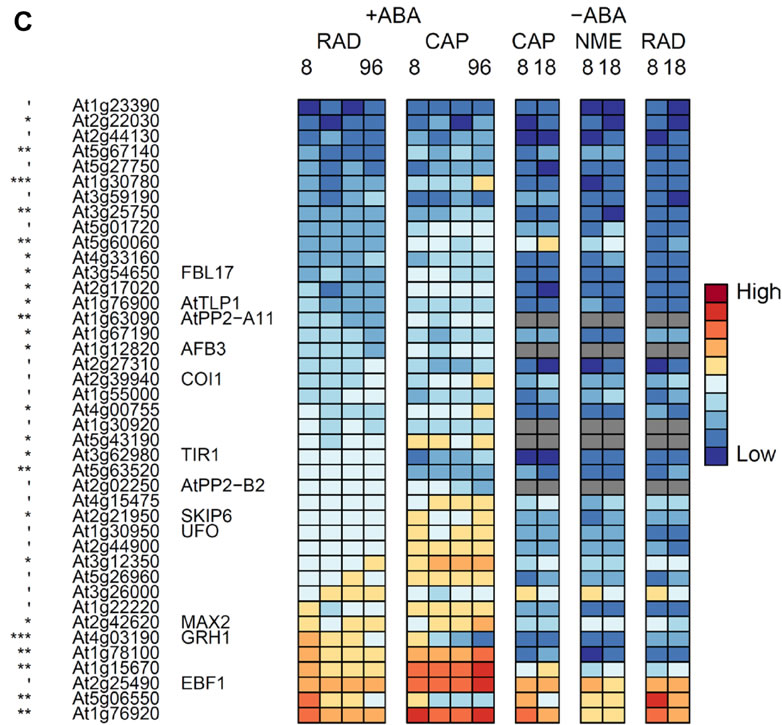
C, Genes encoding F box proteins that are significantly differentially expressed between the RAD and CAP.
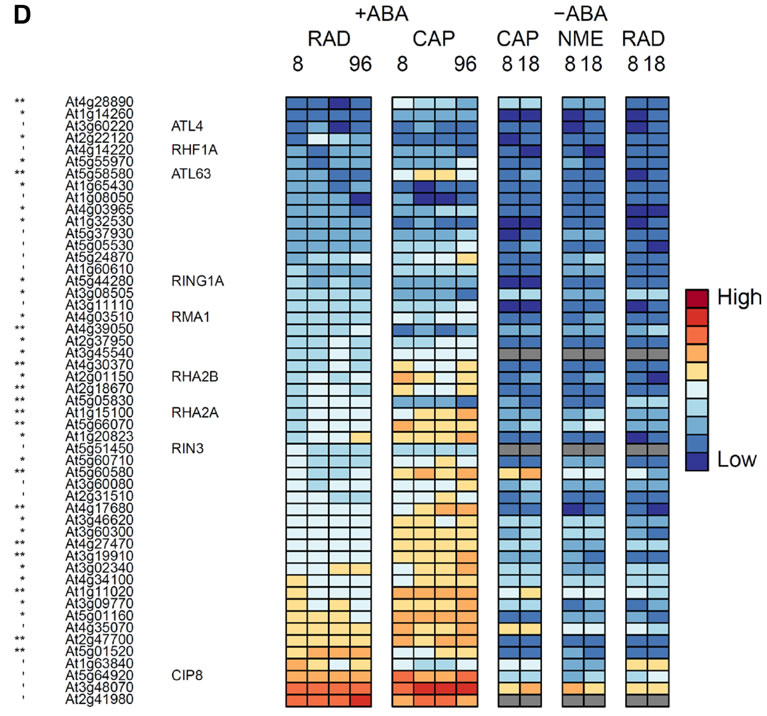
D, Genes encoding RING finger E3 ligaseproteins that are significantly differentially expressed between the RAD and CAP .
', *, **, *** indicate that transcript numbers are significantly different between the tissues on +ABA-arrays at P<0.1, <0.05, <0.01, <0.001 respectively. Genes not present in the data sets are coloured grey.
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |