Journal of Experimental Botany 63: 6325-6334 (2012)
Role of a respiratory burst oxidase of Lepidium sativum (cress) seedlings in root development and auxin signalling [W][OA]
Simon Fraser University, Department of Biological Sciences, 8888 University Drive, Burnaby BC V5A 1S6, Canada (KM, ARK)
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany / Plant Physiology, Schänzlestr. 1, D-79104 Freiburg, German (AL, GLM)
School of Biological Sciences, Plant Molecular Science, Royal Holloway, University of London, Egham, Surrey TW20 0EX, UK (GLM);
Web: 'The Seed Biology Place' - www.seedbiology.eu
* equal contributions
Received 20 July 2012; Revised 11 September 2012; Accepted 21 September 2012
Advance Acess publication 23 October 2012
DOI 10.1093/jxb/ers284
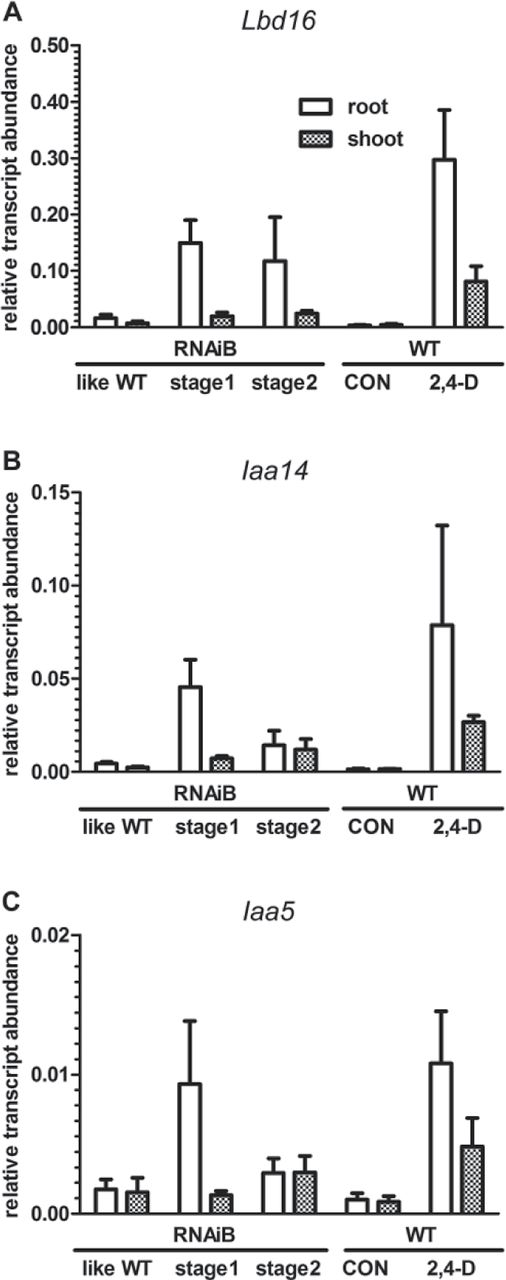
Fig. 5. Expression of auxin-related genes in roots and shoots of L. sativum seedlings of wild-type and Lesarboh-RNAi knockdown lines.
Transcript abundance for the genes encoding the LOBdomain containing protein (LBD) 16 (A), the auxin-responsiveprotein, IAA14 (B) and the auxin-responsive protein IAA5 (C) as determined by qRT-PCR. Stage 1 corresponds to the phenotype shown in Fig. 4B, while stage 2 corresponds to the phenotype shown in Fig. 4C–E. Transcripts in wild-type seedlings were examined under 2,4-D treatment (24 h on 1 μM 2,4-D) and control conditions. Transcript abundance was corrected to the abundance of transcripts associated with the standard genes – the constitutive genes Actin 7 (ACT7; At5g09810) and Elongation Factor 1α (EF1α; At5g60390). Averages of three biological replicates ± SE are presented.
| Article in PDF format (1.8 MB) Supplementary data file (2.3 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |