Journal of Experimental Botany 63: 6325-6334 (2012)
Role of a respiratory burst oxidase of Lepidium sativum (cress) seedlings in root development and auxin signalling [W][OA]
Simon Fraser University, Department of Biological Sciences, 8888 University Drive, Burnaby BC V5A 1S6, Canada (KM, ARK)
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany / Plant Physiology, Schänzlestr. 1, D-79104 Freiburg, German (AL, GLM)
School of Biological Sciences, Plant Molecular Science, Royal Holloway, University of London, Egham, Surrey TW20 0EX, UK (GLM);
Web: 'The Seed Biology Place' - www.seedbiology.eu
* equal contributions
Received 20 July 2012; Revised 11 September 2012; Accepted 21 September 2012
Advance Acess publication 23 October 2012
DOI 10.1093/jxb/ers284
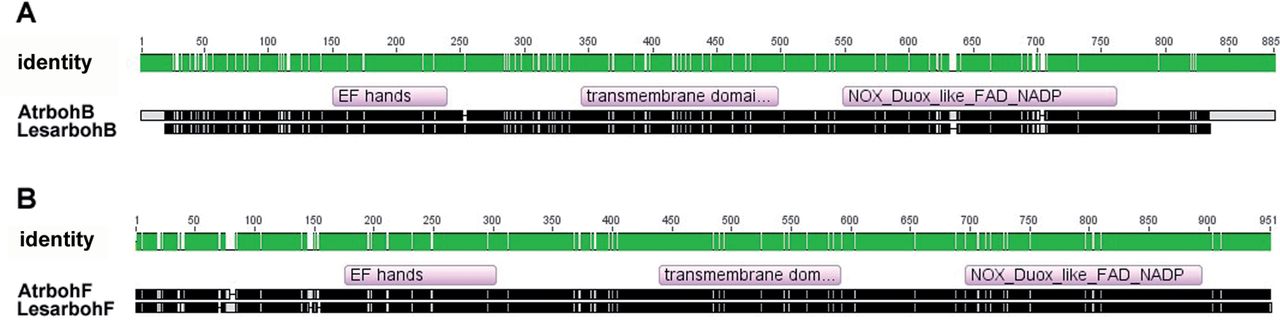
Fig. 1. Alignment of the deduced amino acid sequences of the AtbohB and LesarbohB cDNAs.
(A) Alignment of the AtrbohB amino acid sequence with that of its putative ortholog LesarbohB.
(B) Alignment of the AtrbohF amino acid sequence with that of its putative orthologue LesarbohF. EF hand, transmembrane, and NOX/Duox like FAD/NADP binding domains of RbohB and RbohF of the two plant species were determined with the algorithm DELTA-BLAST (Domain Enhanced Lookup Time Accelerated BLAST) and are indicated in the alignment. Note that the amino acids are missing at the LesarbohB termini due to missing bases in the sequencing. Both the green and black sections in the sequences represent identical amino acids between the two proteins. The top panel with green filling also shows amino acid numbers. For an enlarged alignment detailing the specific amino acid residues, see Supplementary Fig. S1 for RbohB and Supplementary Fig. S2 for RbohF at JXB online.
| Article in PDF format (1.8 MB) Supplementary data file (2.3 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |