Zeitschrift für Naturforschung 49c:
781-790 (1994)
Phenylalanine analogues: Potent inhibitors of phenylalanine ammonia-lyase
are weak inhibitors of phenylalanine-tRNA synthetases
Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues
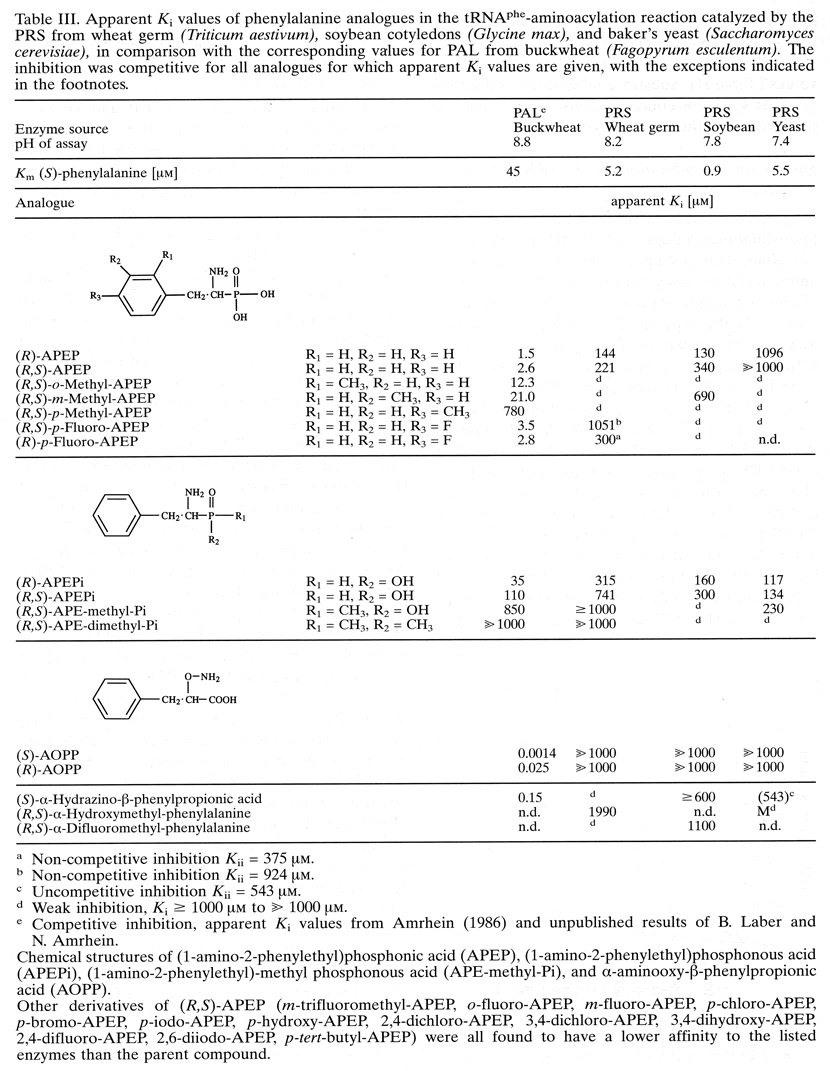
Table III. Apparent Ki values of phenylalanine analogues in the tRNA-phe aminoacylation reaction catalysed by the PRS from wheat germ (Triticum aestivum), soybean cotyledons (Glycine max), and baker's yeast (Saccharomyces cerevisiae), in comparison with the corresponding values for PAL from buckwheat (Fagopyrum esculentum). The inhibition was competitive for those phenylalanine analogues, for which apparent Ki values are given, with the exceptions given at the end of the table.
(a) Non-competitive inhibition Kii = 375 mM.
(b) Non-competitive inhibition Kii = 924 mM.
(c) Uncompetitive inhibition Kii = 543 mM.
(d) Weak inhibition, Ki ≥ 1000 mM to >> 1000 mM.
(e) Competitive inhibition, apparent Ki values from Amrhein (1986) and unpublished results of B. Laber.
Chemical structures of (1-amino-2-phenylethyl)phosphonic acid (APEP), (1-amino-2-phenylethyl)phosphonous acid (APEPi), (1-amino-2-phenylethyl)-methyl phosphonous acid (APE-methyl-Pi), (1-amino-2-phenylethyl)-dimethyl phosphonous acid (APE-dimethyl-Pi), and a-aminooxy-b-phenylpropionic acid (AOPP). Other derivatives of (R,S)-APEP (m-trifluoromethyl-APEP, o-fluoro-APEP, m-fluoro-APEP, p-chloro-APEP, p-bromo-APEP, p-iodo-APEP, p-hydroxy-APEP, 2,4-dichloro-APEP, 3,4-dichloro-APEP, 3,4-dihydroxy-APEP, 2,4-difluoro-APEP, 2,6-diiodo-APEP, p-tert. Butyl-APEP) were all found to have a lower affinity to the listed enzymes than the parent compound.
| Article in PDF format (4.4 MB) | Abstract Fig. 1 Fig. 2 Tab. 1 Tab. 2 Tab. 3 Tab. 4 Phe-analogues |
|
|
The Seed Biology Place
|
Webdesign Gerhard Leubner 2000
|